About LabOUR PUBLICATIONS
The research interests of the Liu Lab include topics in:

Genomics
Transcription regulation

Disease genes prioritization
Machine learning
Lab NewsMore News
LATEST SOFTWARES
Software developed by Liu lab members recently
CrisprCloud
We present a user-friendly, cloud-based, data analysis pipeline for the deconvolution of pooled screening data. This tool serves a dual purpose of extracting, clustering and analyzing raw next generation sequencing files derived from pooled screening experiments while at the same time presenting them in a user-friendly way on a secure web-based platform. Moreover, CRISPRcloud serves as a useful web-based analysis pipeline for reanalysis of pooled CRISPR screening datasets. Taken together, the framework described in this study is expected to accelerate development of web-based bioinformatics tool for handling all studies which include next generation sequencing data.
MARRVEL
MARRVEL (Model organism Aggregated Resources for Rare Variant ExpLoration) aims to facilitate the use of public genetic resources to prioritize rare human gene variants for study in model organisms. To automate the search process and gather all the data in a simple display we extract data from human data bases (OMIM, ExAC, Geno2MP, DGV, and DECIPHER) for efficient variant prioritization. The protein sequences for six organisms (S. cerevisiae, C. elegans, D. melanogaster, D. rerio, M. musculus, and H. sapiens) are aligned with highlighted protein domain information via collaboration with DIOPT. The key biological and genetic features are then extracted from existing model organism databases (SGD, PomBase, WormBase, FlyBase, ZFIN, and MGI).
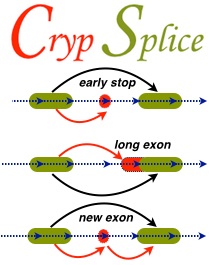
CrypSplice
Alternative splicing of RNA is the key mechanism by which a single gene codes for multiple functionally diverse proteins. Recent studies identified previously unknown class of exons, ‘cryptic’ exons, in RNA transcripts. These cryptic exons are often associated with various human cancers and neurological disorders. Genome-wise detection of cryptic splice sites can facilitate a comprehensive understanding of the underlying disease mechanisms and develop strategies that hope to resolve cryptic splicing with the ultimate goal of therapeutic applications. CrypSplic is a novel cryptic splice site detection method. It uses beta-binomial distribution to model junction count data. Every junction is subjected to a beta binomial test w.r.t conditions and classified to aid molecular inferences.
FIND US
Houston, TX 77030